Functional microbiome analysis
Many microbiome studies require an understanding that goes beyond which microorganisms are present (i.e. the taxonomic composition). Enhanced understandings often come from knowledge about what the microorganisms can do – in other words, their functional potential. Functional microbiome analysis is a method that predicts what microorganisms are equipped to do based on the genes they have, and it provides insights on how they contribute to states of health and disease.
To provide functional insights, we offer functional profiling using Kyoto Encyclopedia of Genes and Genomes (KEGG) and/or curated databases for gut metabolic modules, gut-brain modules, bile-acid metabolism, human milk oligosaccharide (HMO) metabolism, antimicrobial resistance genes (CARD), and more.
Functional Species Groups (FSG)
Different microbial species may occupy the same functional niche in different hosts, a phenomenon known as functional redundancy. Through our Functional Species Group (FSG) tool, we group species together into an FSG if they share the same functional potential regarding metabolic pathways, enzymatic activities, or disease association (e.g. encoding the molecular pathway for producing butyrate via transferase).
Furthermore, a group of functionally related species may be associated with a treatment group without any one of these species being differentially abundant on its own. An enrichment test for FSG can identify such associations (similar to what others call a taxon-set-enrichment analysis), by statistically testing whether multiple species, all belonging to the same FSG, systematically have a stronger association with the treatment group.
The power of this approach is that it identifies functional enrichment in the absence of species or strain enrichment – thus pinpointing functions or pathways of interest for the phenotype and/or treatment.
We are continuously expanding our FSG sets based on new scientific developments and our customers’ interests. We see functional analysis / FSG enrichment analysis as a natural extension of taxonomic analysis in many cases, as it can bring you closer to understanding the underlying mechanism linking the microbiome to the phenotype, disease, and/or treatment response.
Applications
By including functional microbiome analysis in your study, you can:
- Identify potential biomarkers for a phenotype or for disease diagnosis and progression. Functional profiling can be used as input for machine learning in pursuit of these aims.
- Understand adaptation of the microbiome to different environmental conditions, such as toxin exposure, antibiotic use, or changes in diet.
- Inform the development of new treatments by increasing your mechanistic understanding of the phenotype or disease. Such an analysis provides a foundation for generating actionable hypotheses that can be tested in follow-up studies, including in vivo or in vitro experiments. Multi-omics data integration can further enhance your insights.
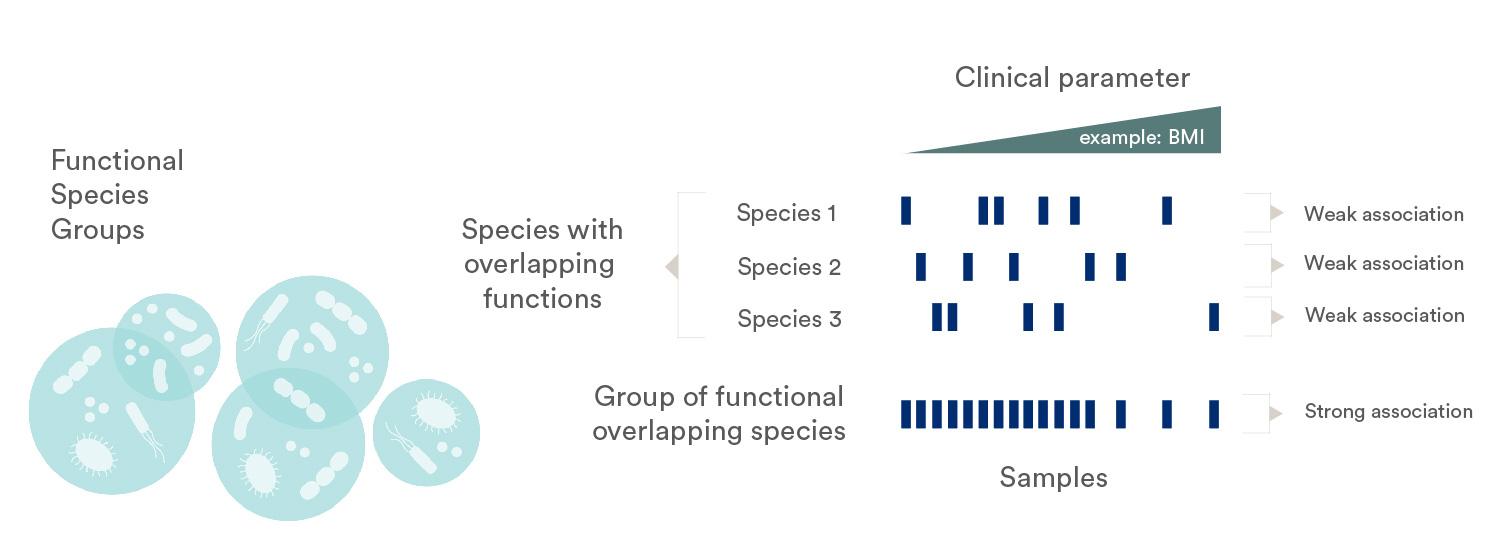
The functional species group concept. Species 1, 2 and 3 share functional potential for KEGG module X, so together these three MGSs comprise the functional species group X. Each of the three species on their own are weakly associated with the clinical parameter shown at the top (here: BMI), and each MGS on its own does not have a signal strong enough to become significant. Yet the three species occupy the same functional niche across different hosts, so when evaluating these three MGS as a functional species group, we can identify the strong association between this functional module and the clinical parameter.
Contact us to learn more about the applicability of functional microbiome analysis to your study, or to receive further details about our methodology.
